This is a VirtualBox VM I made for QIIME 1.9 based on the current version Ubuntu MATE LTS (16.04.3) with some additional improvements.
The developers of QIIME provide a VirtualBox disk image for QIIME 1.9.1. Instructions for using this image is available here. This is an alternative (unofficial) image with the same version of QIIME but with some features and enhancements as described below.
Note
The QIIME developers are recommending that new users start using QIIME 2 rather than QIIME 1 (the version this image includes). I had created the original version of this image when I was using QIIME 1 for my work. This image is only provided with the hope that it will be useful for anyone still requiring QIIME 1.
Features and Enhancements
This image is based on the latest LTS version of Ubuntu 16.04.3 with
Ubuntu MATE - a simpler desktop environment with
minimal applications and desktop effects disabled for better performance.
The default user is qiime with password qiime.
VirtualBox guest additions is pre-installed so features like shared folders, shared clipboard and better video support should all work immediately.
QIIME 1.9.1 and some dependencies (BLAST, Mothur, Clearcut, Infernal,
Muscle, Cdbtools, Swarm, ea-utils, SeqPrep, SortmeRNA, BWA, BLAT) are installed
using the Conda package manager and an environment is setup for the qiime user
called qiime1. This environment is activated automatically on login.
R is installed along with the required packages (ape, biom, optparse, RColorBrewer, randomForest, vegan, DESeq2 and metagenomeSeq).
Additional dependencies installed - RDP Classifier, cd-hit, ChimeraSlayer, rtax, AmpliconNoise, Cytoscape and SourceTracker, thanks to qiime-deploy.
Search and install additional software easily from the Ubuntu and
Debian repositories using the inclued Synaptic Package Manager. Alternatively,
packages can also be installed using the Conda package
manager that is configured for the qiime user. Additional software like
USEARCH can be made available by downloading the executable to
/home/qiime/Software/bin and then creating a symlink or renaming the
executable to the desired name. See tips section below for an example.
Storage space can be increased, if necessary. LVM has been configured for the base installation so hard disk size can be increased if necessary by adding additional disks to the VM in VirtualBox.
Setup and Usage
- Install VirtualBox and the extension pack.
- Download the QIIME_1.9.1_UM.ova file.
- Click on the downloaded file. If that does not launch VirtualBox, open
VirtualBox and then use the
File --> Import Applianceoption and select the downloaded file. - VirtualBox will offer the possibility to configure the number of CPU’s, the amount of RAM and other options in the next step.
Tips
Passwords
Password for the QIIME user qiime is qiime. You can change it using the
passwd command.
Becoming root
The qiime user has admin privileges (sudo) and so logging in to the root
account is not normally required. If you do need to become root, use
sudo su.
How do I exit the qiime1 conda environment?
Use the command source deactivate or dt (alias).
Increasing storage space
- Shutdown the VM.
- VirtualBox -> Settings - Storage -> Click on the “Adds Harddisk” button next to Controller: SATA. From there, select “Create new disk”, use defaults and select a suitable disk size depending on the space available on the hard disk. Click OK when done.
- Start VM, open terminal, use command
lsblkto check if the new hard disk is listed. - Use LVM commands to extend and resize the file system:
pvcreate /dev/sdb
vgextend ubuntu-mate-vg /dev/sdb
lvresize -L +10G -r /dev/ubuntu-mate-vg/root
The root (/) filesystem should have now been resized. To check disk space
usage, use the command df -h.
Testing
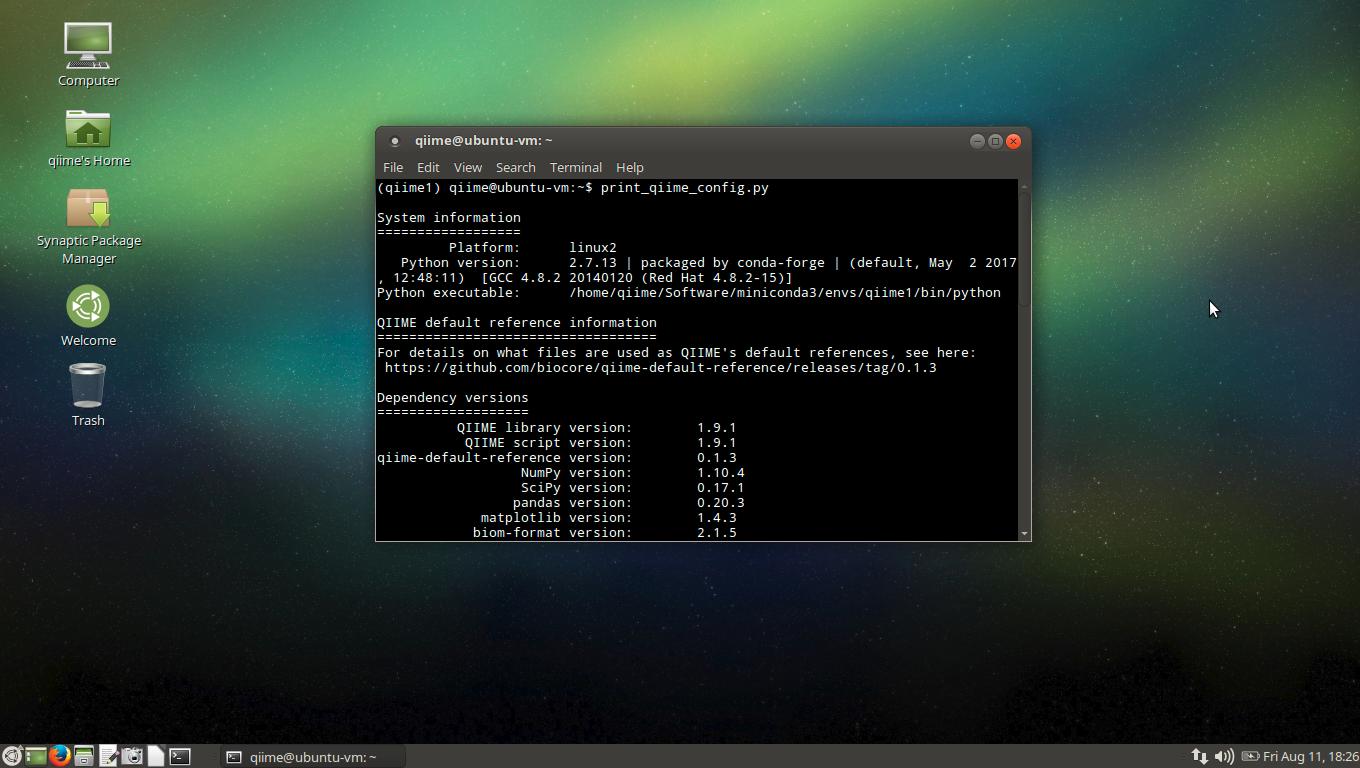
Here is the output of the print_qiime_config.py command to test
the QIIME installation:
(qiime1) qiime@ubuntu-vm:~$ print_qiime_config.py -tvf
System information
==================
Platform: linux2
Python version: 2.7.13 | packaged by conda-forge | (default, May 2 2017, 12:48:11) [GCC 4.8.2 20140120 (Red Hat 4.8.2-15)]
Python executable: /home/qiime/Software/miniconda3/envs/qiime1/bin/python
QIIME default reference information
===================================
For details on what files are used as QIIME's default references, see here:
https://github.com/biocore/qiime-default-reference/releases/tag/0.1.3
Dependency versions
===================
QIIME library version: 1.9.1
QIIME script version: 1.9.1
qiime-default-reference version: 0.1.3
NumPy version: 1.10.4
SciPy version: 0.17.1
pandas version: 0.20.3
matplotlib version: 1.4.3
biom-format version: 2.1.5
h5py version: 2.6.0 (HDF5 version: 1.8.17)
qcli version: 0.1.1
pyqi version: 0.3.2
scikit-bio version: 0.2.3
PyNAST version: 1.2.2
Emperor version: 0.9.51
burrito version: 0.9.1
burrito-fillings version: 0.1.1
sortmerna version: SortMeRNA version 2.0, 29/11/2014
sumaclust version: SUMACLUST Version 1.0.00
swarm version: Swarm 1.2.19 [Jun 1 2017 11:47:02]
gdata: Installed.
RDP Classifier version (if installed): rdp_classifier-2.2.jar
Java version (if installed): Not installed.
QIIME config values
===================
For definitions of these settings and to learn how to configure QIIME, see here:
http://qiime.org/install/qiime_config.html
http://qiime.org/tutorials/parallel_qiime.html
blastmat_dir: None
pick_otus_reference_seqs_fp: /home/qiime/Software/miniconda3/envs/qiime1/lib/python2.7/site-packages/qiime_default_reference/gg_13_8_otus/rep_set/97_otus.fasta
sc_queue: all.q
topiaryexplorer_project_dir: None
pynast_template_alignment_fp: /home/qiime/Software/miniconda3/envs/qiime1/lib/python2.7/site-packages/qiime_default_reference/gg_13_8_otus/rep_set_aligned/85_otus.pynast.fasta
cluster_jobs_fp: start_parallel_jobs.py
pynast_template_alignment_blastdb: None
assign_taxonomy_reference_seqs_fp: /home/qiime/Software/miniconda3/envs/qiime1/lib/python2.7/site-packages/qiime_default_reference/gg_13_8_otus/rep_set/97_otus.fasta
torque_queue: friendlyq
jobs_to_start: 1
slurm_time: None
denoiser_min_per_core: 50
assign_taxonomy_id_to_taxonomy_fp: /home/qiime/Software/miniconda3/envs/qiime1/lib/python2.7/site-packages/qiime_default_reference/gg_13_8_otus/taxonomy/97_otu_taxonomy.txt
temp_dir: /tmp/
slurm_memory: None
slurm_queue: None
blastall_fp: blastall
seconds_to_sleep: 1
QIIME full install test results
===============================
test_FastTree_supported_version (__main__.QIIMEDependencyFull)
FastTree is in path and version is supported ... ok
test_INFERNAL_supported_version (__main__.QIIMEDependencyFull)
INFERNAL is in path and version is supported ... ok
test_R_supported_version (__main__.QIIMEDependencyFull)
R is in path and version is supported ... ok
test_ampliconnoise_install (__main__.QIIMEDependencyFull)
AmpliconNoise install looks sane. ... ok
test_blast_supported_version (__main__.QIIMEDependencyFull)
blast is in path and version is supported ... ok
test_blastall_fp (__main__.QIIMEDependencyFull)
blastall_fp is set to a valid path ... ok
test_blastmat_dir (__main__.QIIMEDependencyFull)
blastmat_dir is set to a valid path. ... ok
test_cdbtools_supported_version (__main__.QIIMEDependencyFull)
cdbtools is in path and version is supported ... ok
test_cdhit_supported_version (__main__.QIIMEDependencyFull)
cd-hit is in path and version is supported ... ok
test_chimeraSlayer_install (__main__.QIIMEDependencyFull)
no obvious problems with ChimeraSlayer install ... ok
test_clearcut_supported_version (__main__.QIIMEDependencyFull)
clearcut is in path and version is supported ... ok
test_cluster_jobs_fp (__main__.QIIMEDependencyFull)
cluster_jobs_fp is set to a valid path and is executable ... ok
test_denoiser_supported_version (__main__.QIIMEDependencyFull)
denoiser aligner is ready to use ... ok
test_for_unrecognized_values (__main__.QIIMEDependencyFull)
qiime_config has no extra values ... ok
test_gdata_install (__main__.QIIMEDependencyFull)
gdata is installed ... ok
test_get_qiime_scripts_dir (__main__.QIIMEDependencyFull)
Test that we can find the directory containing QIIME scripts. ... ok
test_h5py (__main__.QIIMEDependencyFull)
h5py is installed ... ok
test_mothur_supported_version (__main__.QIIMEDependencyFull)
mothur is in path and version is supported ... ok
test_muscle_supported_version (__main__.QIIMEDependencyFull)
muscle is in path and version is supported ... ok
test_pynast_template_alignment_blastdb_fp (__main__.QIIMEDependencyFull)
pynast_template_alignment_blastdb, if set, is set to a valid path ... ok
test_pynast_template_alignment_fp (__main__.QIIMEDependencyFull)
pynast_template_alignment, if set, is set to a valid path ... ok
test_raxmlHPC_supported_version (__main__.QIIMEDependencyFull)
raxmlHPC is in path and version is supported ... ok
test_rtax_supported_version (__main__.QIIMEDependencyFull)
rtax is in path and version is supported ... ok
test_sourcetracker_installed (__main__.QIIMEDependencyFull)
sourcetracker is installed ... ok
test_temp_dir (__main__.QIIMEDependencyFull)
temp_dir is set to a valid path ... ok
test_uclust_supported_version (__main__.QIIMEDependencyFull)
uclust is in path and version is supported ... ok
test_usearch_supported_version (__main__.QIIMEDependencyFull)
usearch is in path and version is supported ... ok
----------------------------------------------------------------------
Ran 27 tests in 0.276s
OK

Comments