My software contributions in research groups, and some side projects.
Biomint Desktop
A ready-to-use Linux virtual machine for Bioinformatics
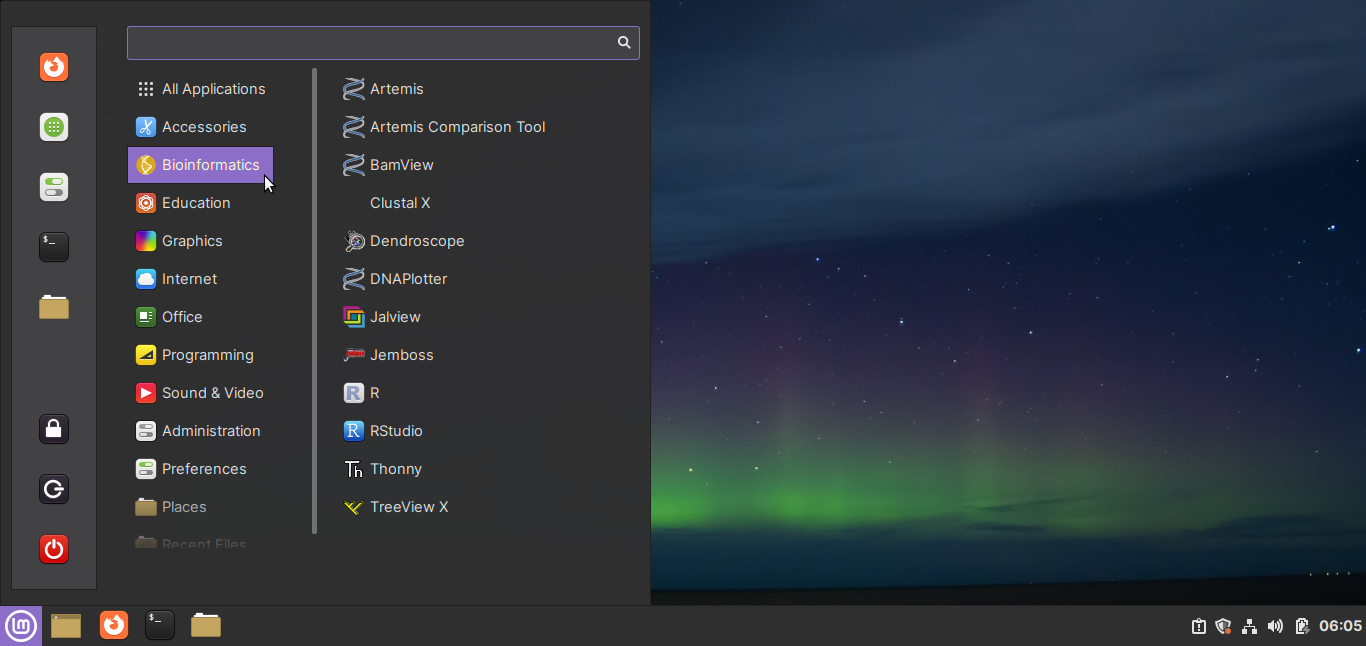
More details will be available shortly!
Note: Biomint was previously known as BioinfoLinux/BioVM.
Archived projects
These are projects I had worked on previously and are not being developed or maintained at present. Source code is available on my GitLab or GitHub accounts.
Docker images
I have built Docker images for the following software when required in the research group or as a learning exercise — VIGA, QIIME 1 1, PyFBA, and DADA 2.
These images include programs and their dependencies pre-installed. In some cases, there are wrapper scripts in Bash, that will help users run containers easily, without needing root privileges. You can get these scripts and Dockerfiles from GitLab.
Related blog post: Docker images for Bioinformatics software
The following projects were developed in research groups as part of a team. This includes RiboPlot, riboSeqR wrapper for Galaxy, IT Query and IT CRISPR. These were all written in Python 2 and are not currently being developed. Python 2 is not supported as of January 2020.
RiboPlot
A Python program to plot and output Ribo-Seq read counts from BAM format alignment files.
This program and the riboSeqR Galaxy wrapper discussed below, were developed under the supervision of Dr. Audrey Michel and Dr. Pasha Baranov at LAPTI, UCC in 2015.
The main program is written in Python 2 and uses the pysam module for working with FASTA and BAM format alignment files. Matplotlib is used for generating plots. HTML output is generated using the Skeleton CSS framework and the Tablesorter JavaScript library is used for sorting the HTML table produced from Ribocount.
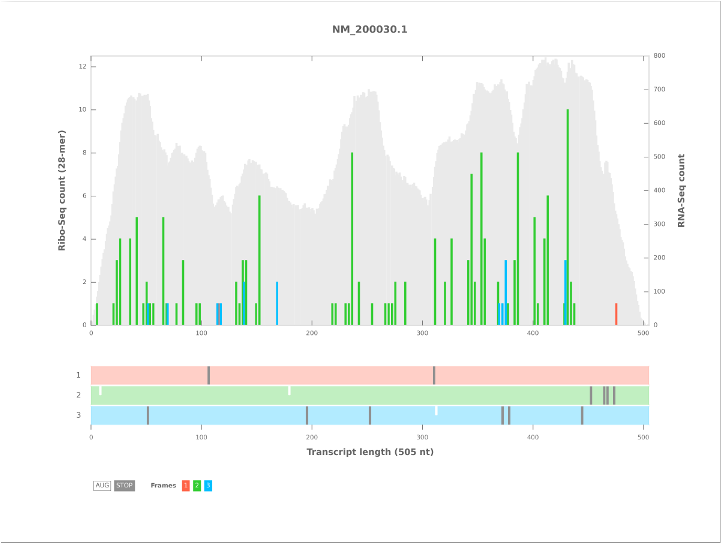
You can get RiboPlot from:
PyPI •
Anaconda •
Galaxy tool shed •
GitHub
Documentation is available here.
Galaxy wrapper for riboSeqR
A web interface (XML wrapper) for integrating the riboSeqR R package in RiboGalaxy or Galaxy.
Using the inclued tools, you can prepare input files for riboSeqR from SAM format alignment files, calculate triplet periodicity, perform metagene analysis, plot ribosome profiles, and perform differential translation analysis using baySeq.
You can install riboseqr_wrapper from Galaxy tool shed.
Source code is available on GitHub.
BioNumerics 6 GUIs and scripts
These programs were written using Python 2 and PyQt 4 and were developed in collaboration with members in the Microbial Phylogeography group at Environmental Research Institute, UCC during 2007-2012 under the supervision of Prof. Mark Achtman.
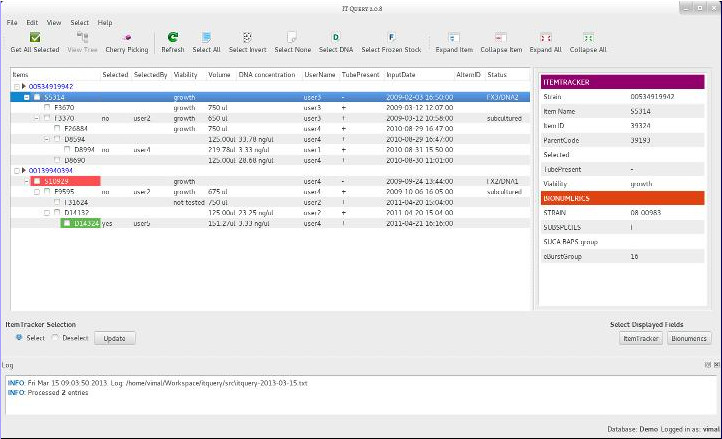
You can get these programs and scripts from GitLab.
GWIPS tools
A collection of Python scripts to download genome annotations and reference sequences from UCSC. You can get them from GitHub.
VLinux
A live Linux CD with Bioinformatics software pre-installed.
Note: There are no new releases of VLinux. Ideas from the project are now incorporated into Biomint.
Themes for Pelican and Sphinx
Themes for Pelican static site generator and Sphinx documentation generator.
Attila-X
A WIP Pelican theme 3 based on Ghost Attila theme.
Susty Pelican
The Wordpress susty 2 theme, ported to Pelican (GitHub).
Solar theme
Sphinx theme using the Solarized colour scheme (GitLab).
-
susty theme for Wordpress. ↩
-
This is the theme used on this website. ↩